4973. Mutation
Genetic scientists from the planet Olympia are once again
conducting experiments with the DNA of primitive organisms. The genome of an
organism is a sequence of genes, each of which can be encoded by a natural
number. Genes encoded by the same number are considered identical, while genes
encoded by different numbers are considered different.
The scientists have already created a primitive organism and
want to modify its genome in such a way as to produce an ideal organism. They
believe that this will help develop cures for many diseases in the future.
An organism is considered ideal if any two identical genes
are either located on adjacent positions in the genome or have at least one
identical gene between them.
In one operation, scientists can select one or more identical
genes in the genome, remove them, and then place them back into the genome,
possibly at different positions. Since each such operation weakens the
organism, the scientists aim to minimize the number of operations needed to achieve
their goal.
Write a program that, given a representation of the genome,
determines the minimum number of operations required to create an ideal
organism.
Input. The first line contains the number of genes n (1 ≤ n ≤ 105) in the genome of a primitive organism. The
second line contains n positive integers, each of which does not exceed n
– the sequence of genes in the genome.
Output. Print the minimum number
of operations required to create an ideal organism.
|
Sample
input |
Sample
output |
|
9 1 2 1 3 1
3 2 4 5 |
2 |
SOLUTION
greedy
Algorithm analysis
The gene
rearrangement should be performed in such a way that identical genes are placed
next to each other. At the same time, it is necessary to minimize the number of
operations performed.
For each positive integer ai, we associate an interval [si; ei], where si is the position where ai first appears, and ei is the position where ai appears for the
last time.
Let res
be the maximum possible number of non-overlapping intervals. This value can be
found using the “activity-selection” algorithm, which means that each of
the remaining intervals overlaps with at least one of the selected intervals.
After this, the described procedure should be applied to the genes
corresponding to these intervals.
If k is the total
number of constructed intervals, then the answer to the problem will be k – res.
Example
For the given
example, the
intervals will look like this (position numbering starts from zero):
·
for 1: [0; 4];
·
for 2: [1; 6];
·
for 3: [3; 5];
·
for 4: [7; 7];
·
for 5: [8; 8];
Let’s sort the intervals
by their end coordinate:
[0; 4], [3; 5], [1; 6], [7; 7], [8; 8]
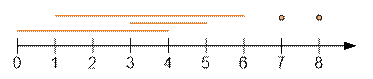
The maximum number of
non-overlapping intervals is 3. For example, we can select the intervals [0; 4], [7; 7],
[8; 8]. For
the genes corresponding to the remaining intervals, the procedure
described in the problem statement should be applied. The corresponding genes
are 2 and 3. Two rearrangements can be performed, for example, as follows:
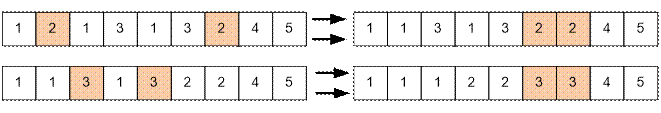
Algorithm implementation
Let MAX be the maximum possible number of genomes in the
input sequence.
#define MAX 100001
The vector of pairs v stores the segments [si;
ei].
vector<pair<int, int> > v;
int s[MAX], e[MAX];
Read the input data. Initialize s[i] =
-1, e[i] = -1.
scanf("%d", &n);
memset(s, -1, sizeof(s));
memset(e, -1, sizeof(e));
for (i = 0; i < n; i++)
{
scanf("%d", &x);
If the number x appears for the first
time, record its position in the input sequence in s[x].
if (s[x] == -1) s[x] = i;
If the number x does not appear for the first time, record its
position in e[x], assuming that x
appears for the last time.
if (i > e[x]) e[x] = i;
}
All the constructed intervals are stored in the vector v.
for (i = 1; i <= n; i++) //
numbers 1..n
if (s[i] != -1) v.push_back(make_pair(e[i], s[i]));
The intervals are stored in the vector in reverse order (in the form [ei; si]) so that, by default, the intervals are sorted in
ascending order of their ends.
sort(v.begin(), v.end());
Find the maximum number res
of non-overlapping
intervals.
i = res =
0;
while (i < v.size())
{
Add the i-th interval to the set
of non-overlapping intervals. The variable temp holds the end of this
interval (let’s call it the current interval).
res++;
temp = v[i++].first; // end
As long as the start of the next interval is less than the end of the
current interval, skip such an interval, as it overlaps with the current one.
while (i < v.size() && v[i].second < temp) i++;
}
Print the answer.
printf("%d\n", v.size() -
res);